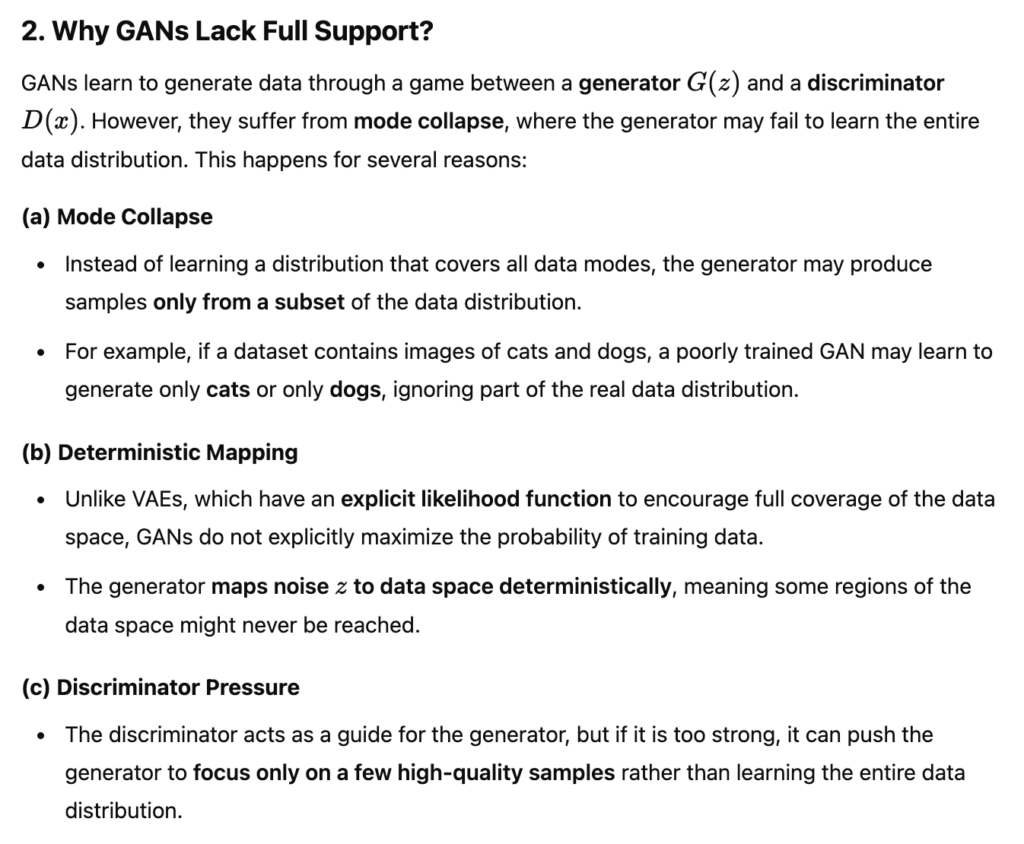
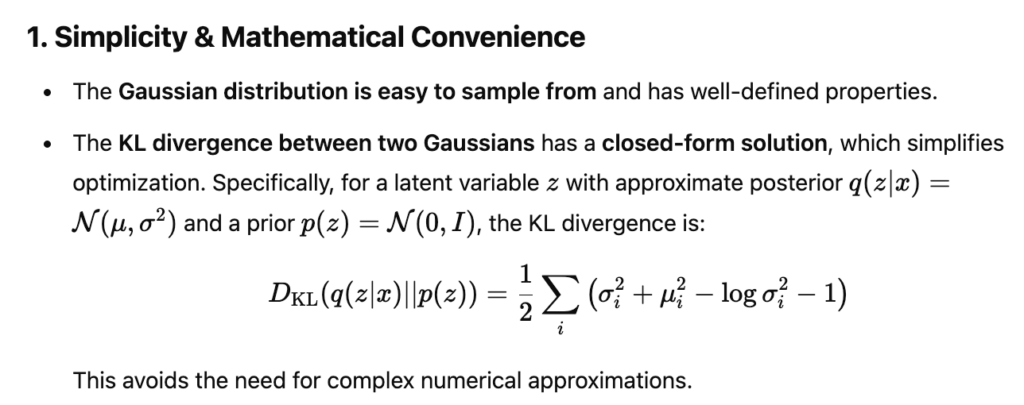
For an experiment of metaformer, I was trying to add CIFAR100 dataset into the training script. Since CIFAR100 is too small, I need to let it repeat mulitple times in one epoch. Therefore I add a new type of dataset:
class RepeatDataset(Dataset):
def __init__(self, dataset, repeats):
self.dataset = dataset
self.repeats = repeats
self.length = len(dataset) * repeats
def __getitem__(self, idx):
return self.dataset[idx % len(self.dataset)]
def __len__(self):
return self.lengthBut the training will report error:
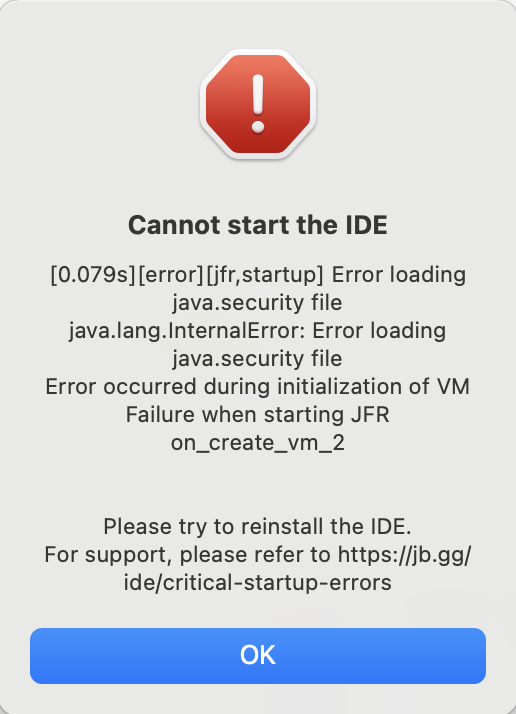
Traceback (most recent call last):
File "/home/robin/code/metaformer/train.py", line 970, in <module>
main()
File "/home/robin/code/metaformer/train.py", line 732, in main
train_metrics = train_one_epoch(
^^^^^^^^^^^^^^^^
File "/home/robin/code/metaformer/train.py", line 798, in train_one_epoch
for batch_idx, (input, target) in enumerate(loader):
^^^^^^^^^^^^^^^^^
File "/home/robin/miniconda3/envs/poolformer/lib/python3.12/site-packages/timm/data/loader.py", line 131, in __iter__
for next_input, next_target in self.loader:
^^^^^^^^^^^
File "/home/robin/miniconda3/envs/poolformer/lib/python3.12/site-packages/torch/utils/data/dataloader.py", line 733, in __next__
data = self._next_data()
^^^^^^^^^^^^^^^^^
File "/home/robin/miniconda3/envs/poolformer/lib/python3.12/site-packages/torch/utils/data/dataloader.py", line 1515, in _next_data
return self._process_data(data, worker_id)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/robin/miniconda3/envs/poolformer/lib/python3.12/site-packages/torch/utils/data/dataloader.py", line 1550, in _process_data
data.reraise()
File "/home/robin/miniconda3/envs/poolformer/lib/python3.12/site-packages/torch/_utils.py", line 750, in reraise
raise exception
AttributeError: Caught AttributeError in DataLoader worker process 0.
Original Traceback (most recent call last):
File "/home/robin/miniconda3/envs/poolformer/lib/python3.12/site-packages/torch/utils/data/_utils/worker.py", line 349, in _worker_loop
data = fetcher.fetch(index) # type: ignore[possibly-undefined]
^^^^^^^^^^^^^^^^^^^^
File "/home/robin/miniconda3/envs/poolformer/lib/python3.12/site-packages/torch/utils/data/_utils/fetch.py", line 55, in fetch
return self.collate_fn(data)
^^^^^^^^^^^^^^^^^^^^^
File "/home/robin/miniconda3/envs/poolformer/lib/python3.12/site-packages/timm/data/mixup.py", line 305, in __call__
output = torch.zeros((batch_size, *batch[0][0].shape), dtype=torch.uint8)
^^^^^^^^^^^^^^^^^
AttributeError: 'Image' object has no attribute 'shape'. Did you mean: 'save'? It cost me a quite long time to solve it. The key is in the implementation of “timm.data.create_loader”: https://github.com/huggingface/pytorch-image-models/blob/main/timm/data/loader.py#L291. In it, it will set “dataset.transform” to a new value, and in “timm.data.dataset” https://github.com/huggingface/pytorch-image-models/blob/main/timm/data/dataset.py#L66-L67, it will check and use this new set “transform”:
...
if self.transform is not None:
img = self.transform(img)
...Since the class RepeatDataset is created by myself and it will not handle the “dataset.transform = create_transform()”, it failed when calling the non-existed “transform()”.
The fix comes from ChatGPT and I think it’s not bad:
class RepeatDataset(Dataset):
def __init__(self, dataset, repeats):
self.dataset = dataset
self.repeats = repeats
self.length = len(dataset) * repeats
@property
def transform(self):
return self.dataset.transform
@transform.setter
def transform(self, value):
self.dataset.transform = value
def __getitem__(self, idx):
return self.dataset[idx % len(self.dataset)]
def __len__(self):
return self.length